The Gut-Centric Pathogenesis of Long COVID: A Synthesis of Mechanistic and Clinical Evidence Linking SARS-CoV-2, Microbiome Dysbiosis, and Post-Acute Sequelae
Persistent SARS-CoV-2 infection in the gut drives microbiome dysbiosis, barrier failure, and systemic inflammation, forming a core mechanism of Long COVID pathogenesis.
Executive Summary
The global health crisis precipitated by the SARS-CoV-2 pandemic has extended far beyond the acute phase of COVID-19, leaving millions to contend with a chronic, multisystemic condition known as Post-Acute Sequelae of COVID-19 (PASC), or Long COVID. While initially understood as a primary respiratory illness, a vast and growing body of scientific evidence now points to the gastrointestinal (GI) tract as a central nexus in the pathophysiology of Long COVID. This report synthesizes findings from mechanistic laboratory studies and observational human cohorts to construct a unified, gut-centric model of Long COVID pathogenesis.
Mechanistic evidence from in vitro and preclinical models provides definitive proof that SARS-CoV-2 can productively infect human intestinal enterocytes, which express the highest levels of the viral entry receptor ACE2 in the body. This enteric infection is not benign; it triggers local inflammation, damages the intestinal barrier, and induces a state of gut microbiome dysbiosis, a process confirmed in animal models where respiratory infection leads to secondary gut pathology.
Observational human studies corroborate and expand upon these findings. Patients with acute COVID-19 exhibit a characteristic gut dysbiosis, marked by a depletion of beneficial, anti-inflammatory bacteria and an enrichment of opportunistic pathogens, with the severity of this imbalance correlating with acute disease severity. Crucially, evidence demonstrates that SARS-CoV-2 can establish a persistent reservoir in the gut, with viral antigens and RNA detected in GI tissue for months to years post-infection. In individuals who develop Long COVID, the acute dysbiosis fails to resolve, instead evolving into a chronic state that is predictive of and correlated with specific PASC symptoms.
The convergence of this evidence points to a core pathophysiological cascade. Persistent viral antigens in the gut are hypothesized to induce overexpression of zonulin, a protein that regulates intestinal tight junctions. This leads to intestinal barrier failure—a "leaky gut"—allowing the translocation of viral components (e.g., spike protein) and microbial products (e.g., lipopolysaccharide) into the systemic circulation. This constant leakage fuels chronic, low-grade systemic inflammation. Concurrently, the dysbiotic microbiome produces an altered profile of metabolites, most notably a deficit in the anti-inflammatory short-chain fatty acid butyrate and a disruption of tryptophan metabolism, which impairs serotonin production and vagus nerve function. This cascade of gut-derived inflammation, immune dysregulation, and metabolic disruption, communicated systemically and via the microbiota-gut-brain axis, offers a compelling explanation for the diverse, multisystemic symptoms of Long COVID, from fatigue and GI distress to cognitive "brain fog" and neurological deficits.
Finally, this gut-centric model opens new avenues for diagnostics and therapeutics. Distinct microbial and metabolic signatures hold promise as objective biomarkers for Long COVID. Furthermore, emerging clinical trials targeting the gut—using synbiotics to restore microbial balance and zonulin antagonists to heal the gut barrier—have shown initial success in alleviating Long COVID symptoms, validating the gut as a critical therapeutic target. This report provides a comprehensive foundation for understanding this complex post-viral syndrome and charting a course toward effective, evidence-based interventions.
Introduction: The Gastrointestinal Tract as a Central Player in Systemic Post-Viral Illness
The emergence of SARS-CoV-2 presented an unprecedented challenge to global health, primarily characterized by acute respiratory distress. However, as the pandemic has evolved, it has become alarmingly clear that for a substantial subset of individuals, the resolution of acute symptoms does not signify recovery. Instead, they experience a constellation of persistent, debilitating symptoms collectively termed Post-Acute Sequelae of COVID-19 (PASC), or Long COVID.1 This syndrome can affect nearly every organ system, with common manifestations including profound fatigue, cognitive dysfunction ("brain fog"), post-exertional malaise, cardiovascular issues, and gastrointestinal disturbances.1 The sheer scale of this chronic illness has catalyzed an urgent scientific quest to understand its underlying biological mechanisms.
In this quest, the gastrointestinal (GI) tract has emerged from a peripheral site of occasional acute symptoms to a central stage in the unfolding drama of Long COVID. The recognition of bidirectional communication pathways, such as the gut-lung axis and the microbiota-gut-brain axis, has provided a critical framework for understanding how a localized event in the gut can have profound systemic consequences.2 The gut-lung axis posits that microbial and immune signals from the gut can modulate respiratory immunity, and vice versa, linking respiratory infections to alterations in the gut microbiome.3 The microbiota-gut-brain axis describes the intricate network of neural, endocrine, and immune signaling through which the gut microbiome influences brain function, mood, and cognition.2 SARS-CoV-2 infection appears to profoundly disrupt these homeostatic axes, initiating a cascade of pathology that persists long after the virus is cleared from the respiratory tract.
This report advances a central thesis: that the pathogenesis of Long COVID is, in many individuals, driven by a sequence of events originating in the gastrointestinal tract. This sequence begins with the direct infection of intestinal cells by SARS-CoV-2, which establishes a persistent viral reservoir. This reservoir, in turn, perpetuates local inflammation and drives a chronic state of gut microbiome dysbiosis. The combination of viral persistence and dysbiosis leads to a critical failure of the intestinal barrier—a "leaky gut"—allowing viral antigens and microbial components to translocate into the systemic circulation. This leakage fuels the chronic, low-grade inflammation and immune dysregulation that characterize Long COVID. Furthermore, the altered microbiome produces a dysfunctional metabolic output, disrupting key signaling pathways that regulate systemic immunity and neurological function. By synthesizing evidence from two distinct but complementary lines of inquiry—mechanistic laboratory studies and observational human cohort studies—this report will build a comprehensive, evidence-based model of the gut-centric pathophysiology of Long COVID, illuminating a path toward targeted diagnostics and therapeutics.
Part I: Mechanistic Evidence of SARS-CoV-2 and Gut Interaction
Before examining the complex clinical picture in human patients, it is essential to establish the biological plausibility of the gut's role in COVID-19 and its sequelae. Mechanistic studies using controlled laboratory models—from cell cultures and organoids to preclinical animal models—provide the foundational evidence that SARS-CoV-2 can directly interact with the intestinal environment and initiate a cascade of pathological events. This section dissects that evidence, demonstrating not only that the virus can infect the gut but that this infection has direct, measurable consequences on intestinal cell biology, barrier function, and local immunity.
Section 1.1: Viral Tropism and Replication in the Intestinal Milieu
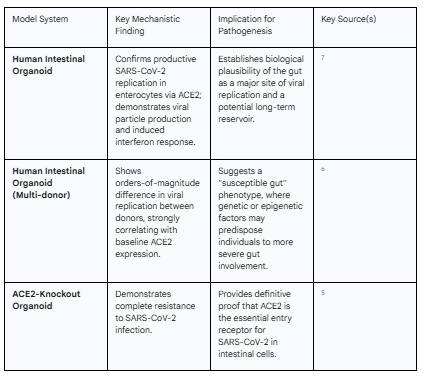
A fundamental prerequisite for the gut-centric hypothesis of Long COVID is demonstrating that SARS-CoV-2 can effectively infect and replicate within intestinal cells. A confluence of evidence from in vitro and ex vivo models has provided definitive proof of this capability, establishing the GI tract as a prime target for the virus.
The tropism of SARS-CoV-2 for the gut is rooted in molecular biology. Viral entry into host cells is mediated by the binding of the viral spike (S) protein to the angiotensin-converting enzyme 2 (ACE2) receptor, followed by priming by cellular proteases, most notably transmembrane serine protease 2 (TMPRSS2).1 Strikingly, human intestinal enterocytes, particularly in the small intestine, express the highest levels of ACE2 in the entire human body, and they co-express TMPRSS2.5 This molecular profile makes the intestinal lining an exceptionally permissive environment for SARS-CoV-2 infection.
This biological potential has been unequivocally confirmed using human intestinal organoids. These are sophisticated three-dimensional cell cultures derived from primary human tissue that self-organize to mimic the architecture and cellular diversity of the intestinal epithelium, providing a compelling model for studying host-pathogen interactions.7 Multiple independent research groups have demonstrated that when SARS-CoV-2 is introduced to these organoids, the virus rapidly infects enterocytes.7 Using advanced techniques like transmission electron microscopy, scientists have visualized the entire viral lifecycle within these cells, from the formation of double-membrane vesicles for replication to the assembly and release of new, mature virus particles.7 This confirms that the infection is not abortive but productive, meaning the gut can serve as a factory for viral propagation.7 Furthermore, RNA sequencing of infected organoids reveals the activation of interferon-stimulated genes, indicating that the intestinal cells recognize the virus and mount a characteristic antiviral response.7 The essential role of ACE2 in this process was elegantly proven in studies using ACE2-knockout intestinal organoids; these genetically modified organoids were found to be completely resistant to SARS-CoV-2 infection, cementing ACE2 as the critical entry gateway in the gut.5
Beyond simply confirming infectivity, organoid studies have uncovered a critical layer of complexity: significant inter-individual variability. When intestinal organoid-derived monolayers from different human donors were infected, they displayed orders-of-magnitude differences in the levels of viral replication.6 This dramatic variation in susceptibility was not random. It correlated strongly and directly with the baseline expression level of ACE2 in the donor's original tissue.6 Factors such as the donor's age, sex, or underlying inflammatory bowel disease (IBD) status did not account for this difference.6 This finding has profound implications, suggesting the existence of a "susceptible gut" phenotype. The reasoning follows a clear path: an individual's genetically or epigenetically determined level of intestinal ACE2 expression may dictate the initial magnitude of enteric infection. Higher ACE2 expression allows for more efficient viral entry, leading to a larger burst of replication and a greater number of initially infected cells. This larger initial viral burden in the gut could then establish a more robust and difficult-to-clear viral reservoir. Therefore, baseline intestinal ACE2 expression may function as a pre-existing, non-modifiable risk factor that predisposes certain individuals to more significant gut involvement and, consequently, a higher likelihood of developing Long COVID. This moves the question from a simple "Can the virus infect the gut?" to a more nuanced "How does the extent of initial gut infection influence long-term outcomes?"
Section 1.2: Pathophysiological Consequences in Preclinical Animal Models
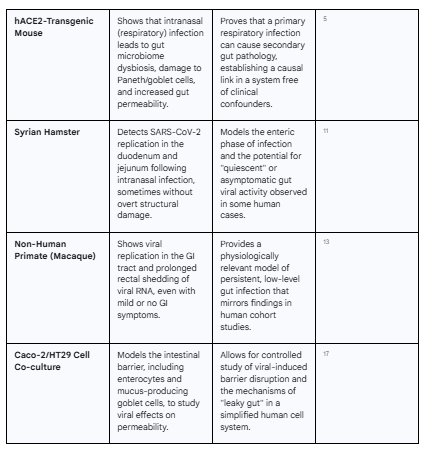
While organoids provide a powerful window into human cellular interactions, preclinical animal models are indispensable for understanding how these interactions translate to whole-organism pathophysiology. They allow researchers to study the interplay between different organ systems and establish causality in ways that are impossible in human studies, which are often confounded by variables like medications and comorbidities.9 Animal models of SARS-CoV-2 infection, though each with limitations, collectively corroborate the findings from organoid studies and demonstrate that a primary respiratory infection can indeed cause significant downstream pathology in the gut.
Murine models have been particularly instrumental. Standard laboratory mice are not naturally susceptible to SARS-CoV-2 due to differences in their ACE2 receptor. However, this has been overcome by using mice genetically engineered to express human ACE2 (hACE2) or by sensitizing their respiratory tracts to the virus.10 In a landmark study, when hACE2-transgenic mice were infected with SARS-CoV-2 via the intranasal route—mimicking a primary respiratory infection—they subsequently developed significant gut microbiome dysbiosis.5 This was accompanied by histological changes in the intestine, including alterations to specialized epithelial cells like Paneth cells (which produce antimicrobial peptides) and goblet cells (which produce protective mucus), as well as an increase in markers of gut barrier permeability.9 This finding is crucial because it establishes a causal link: a respiratory infection can systemically seed the virus, leading to secondary pathology in the GI tract. This supports a model where the virus, after replicating in the lungs, disseminates throughout the body, likely via the bloodstream, and establishes a secondary site of infection in the ACE2-rich gut. This implies that even patients without initial GI symptoms are likely to experience gut involvement, positioning the gut as a key site for viral persistence long after the respiratory infection has resolved.
Hamster models, particularly the Syrian hamster, have proven to be robust models for COVID-19 respiratory disease and also show GI involvement.10 Following intranasal infection, SARS-CoV-2 replication has been detected in the duodenum and jejunum of Syrian hamsters.12 While severe structural damage to the gut is not always a feature, the consistent finding of viral replication confirms the gut as a target organ.12 The Roborovski dwarf hamster, which develops a more fulminant disease course, exhibits high viral loads in the jejunum, further linking gut infection to disease severity.12
Non-human primate (NHP) models, such as rhesus and cynomolgus macaques, offer the closest parallel to human physiology. Following infection, these NHPs demonstrate viral replication in the GI tract and can exhibit prolonged rectal shedding of viral RNA, mirroring the observations in human patients.13 This is true even when overt GI symptoms are mild or absent, effectively modeling the "quiescent" or asymptomatic gut infection that has been described in humans.13 Interestingly, post-mortem analyses of infected NHPs sometimes reveal the presence of viral RNA in the gut but no detectable viral antigen, a discrepancy that underscores the challenges in detecting persistent infection and highlights the varying sensitivity of different measurement techniques.13 Collectively, these animal models provide a vital bridge from cell culture to clinical observation, confirming that SARS-CoV-2 targets the gut in vivo and that this enteric involvement can be a direct consequence of an initial respiratory infection.
Section 1.3: The Direct Viral Impact on Gut Barrier and Immune Function
The replication of SARS-CoV-2 within intestinal enterocytes is not a passive event. Mechanistic studies show that it actively triggers a local inflammatory cascade and inflicts damage upon the cellular architecture of the gut lining, initiating the critical process of barrier function breakdown. This local disruption is the first step in the pathway that ultimately leads to systemic disease.
Upon entering an intestinal cell and beginning to replicate, the virus is recognized by the host's innate immune system. This recognition triggers the infected enterocytes to produce and secrete a range of pro-inflammatory signaling molecules, including cytokines like interleukins (ILs) and tumor necrosis factor-alpha (TNF-α), as well as chemokines.5 These molecules act as an alarm system, recruiting various inflammatory immune cells, such as macrophages and lymphocytes, from the circulation into the gut tissue.5 This influx of immune cells creates a localized state of inflammation within the intestinal wall.
This virally-induced inflammation has a profound and immediate impact on the gut microbiome. The altered chemical and cellular environment is less hospitable to many beneficial commensal bacteria and more favorable to opportunistic or pro-inflammatory species, thus directly contributing to the onset of dysbiosis.5 This creates a vicious cycle: the viral infection triggers inflammation, which in turn drives dysbiosis, and the resulting dysbiotic microbiome, often characterized by an overgrowth of pathobionts, further exacerbates the inflammatory state.15
Furthermore, as observed in mouse models, the viral infection and associated inflammation can directly damage the specialized cells that are essential for maintaining the gut's defensive barrier. Alterations to Paneth cells compromise the secretion of antimicrobial peptides, weakening the gut's first line of chemical defense against microbial invaders.9 Damage to goblet cells reduces the production of mucus, thinning the protective physical barrier that separates luminal contents from the intestinal epithelium.9 The combined effect of this cellular damage and persistent inflammation compromises the integrity of the gut barrier, setting the stage for the "leaky gut" phenomenon that is central to the pathogenesis of Long COVID.
Table 1: Summary of Mechanistic Studies on SARS-CoV-2 and the Gut
The following table summarizes the key findings from diverse laboratory models, providing a consolidated view of the mechanistic evidence supporting the gut's role in SARS-CoV-2 infection.
Part II: Observational Evidence from Human Cohort Studies
While mechanistic studies establish what can happen in a controlled setting, observational studies of human patients reveal what is happening during and after natural infection. This section bridges the gap from the laboratory to the clinic, examining the wealth of data from patient cohorts that collectively paint a detailed picture of gut involvement in acute COVID-19 and its evolution into Long COVID. These studies confirm the patterns of dysbiosis, viral persistence, and immune disruption predicted by the mechanistic models and provide critical insights into the long-term consequences.
Section 2.1: The Gut Microbiome Signature of Acute COVID-19
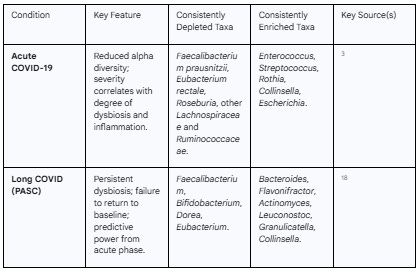
Across the globe, numerous cohort studies involving thousands of patients have converged on a remarkably consistent finding: acute SARS-CoV-2 infection is associated with a profound and characteristic disruption of the gut microbiome, a state known as dysbiosis.15 This is not a subtle shift but a significant alteration of the entire microbial ecosystem.
The most consistent feature of this acute dysbiosis is a marked decrease in overall bacterial diversity.3 A healthy gut microbiome is a rich and diverse community, and this diversity is a cornerstone of its resilience and functionality. In COVID-19 patients, this richness is lost. This loss of diversity is accompanied by a dramatic compositional shift. Specifically, there is a significant depletion of beneficial, commensal bacteria that are known to play a crucial role in maintaining host health.3 Chief among these are the producers of short-chain fatty acids (SCFAs), such as butyrate. Genera like Faecalibacterium, Eubacterium, Roseburia, and other members of the Lachnospiraceae and Ruminococcaceae families, which are powerhouse butyrate producers, are consistently found at reduced levels in COVID-19 patients.3 These bacteria and their metabolic products are vital for nourishing intestinal cells, maintaining the gut barrier, and exerting potent anti-inflammatory effects throughout the body.
As these beneficial microbes decline, a vacuum is created that is filled by an enrichment of opportunistic pathogens and pathobionts—microbes that can cause disease under certain conditions. Studies consistently report an overgrowth of bacteria such as Enterococcus, Streptococcus, Rothia, and Collinsella in the guts of COVID-19 patients.3 The presence of these organisms is often associated with pro-inflammatory activity and has been linked to various chronic inflammatory conditions.
Crucially, the extent of this dysbiosis is not uniform across all patients; it directly correlates with the severity of the acute illness. Patients with more severe COVID-19 exhibit a more profound dysbiosis, which is also associated with higher circulating levels of inflammatory cytokines and markers of tissue damage.3 This suggests a direct link between the state of the gut microbiome and the host's systemic inflammatory response. Furthermore, this microbial signature appears to be specific to SARS-CoV-2, with studies showing that the gut microbiome of COVID-19 patients is distinct from that of patients with other severe respiratory viral infections, such as influenza.3 This unique dysbiotic signature, established during the acute phase of infection, appears to set the stage for the long-term complications that define Long COVID.
Section 2.2: The Persistent Viral Reservoir in the Gastrointestinal Tract
A central and increasingly well-supported tenet of the gut-centric model of Long COVID is that the GI tract serves as a long-term reservoir for SARS-CoV-2. Unlike the respiratory tract, where the virus is typically cleared within weeks, the gut can harbor viral components for months or even years, providing a continuous source of antigenic stimulation that drives chronic immune activation.
Early in the pandemic, it was established that a significant proportion of COVID-19 patients shed viral RNA in their feces. Studies reported that nearly 50% of patients had detectable viral RNA in stool during the acute phase, and this shedding could persist long after respiratory samples turned negative.3 One longitudinal study documented fecal viral RNA shedding for as long as seven months in a subset of patients.3 While the detection of RNA alone does not prove the presence of infectious virus, it was the first clue that the GI tract was a site of prolonged viral presence.
More sophisticated molecular techniques have since provided much stronger evidence for active viral activity in the gut. By analyzing the ratio of different viral RNA fragments, researchers have been able to detect a transcriptional signature indicative of active viral replication in the gut of patients, even in those without GI symptoms and after the virus was no longer detectable in their respiratory tract.14 This discovery of a "quiescent" but metabolically active gut infection was a critical breakthrough, suggesting that the virus was not merely composed of inert fragments but was actively transcribing its genes within the intestinal environment.
The most definitive evidence for a gut reservoir comes from studies that have directly analyzed patient tissue. Researchers have successfully detected SARS-CoV-2 antigens, such as the spike protein, and viral RNA in gut biopsy samples taken from individuals more than a year after their initial infection.19 One study found viral RNA in gut tissue up to two years post-infection.19 Critically, these viral remnants were often found in the connective tissue of the gut wall, in close proximity to resident immune cells, providing a direct anatomical link between the persistent virus and chronic immune stimulation.19 This persistent gut reservoir is now considered a key upstream driver of Long COVID. It is hypothesized to be the source of the persistent inflammation and immune dysregulation that underlie many Long COVID symptoms, including the neurological deficits that may arise from disruption of the serotonin pathway, a process initiated by the inflammatory response to the lingering virus in the gut.20
Section 2.3: Defining the Gut Microbiome of Long COVID
The profound dysbiosis initiated during acute COVID-19 does not necessarily resolve in individuals who go on to develop Long COVID. Instead, longitudinal studies tracking patients over time reveal that the gut microbiome embarks on one of two distinct trajectories. In those who achieve full recovery, the microbiome composition tends to gradually return toward a healthy, pre-infection state. In stark contrast, individuals who develop Long COVID are characterized by a gut microbiome that remains in a distinct, chronic state of dysbiosis.18 This persistent microbial imbalance is not merely a symptom of Long COVID; compelling evidence suggests it is both a predictor and a driver of the condition.
One of the most powerful findings in this field is the predictive capacity of the gut microbiome. Multiple studies have now used machine learning models to analyze microbiome data collected from patients during their acute infection. These models have demonstrated that the composition of the gut microbiome at the time of initial illness can predict with high accuracy which individuals will later develop Long COVID.21 This predictive power is remarkable because it suggests that the gut microbiome is a more sensitive indicator of future risk than standard clinical variables like the severity of the acute illness, age, or comorbidities.21 This finding fundamentally shifts the perspective on the microbiome's role. It implies that the gut microbiome is not just reacting to the chronic disease state of Long COVID but is a key determinant in its development.
This observation has led to a more refined understanding of host resilience. The ability of the acute-phase microbiome to predict long-term outcomes suggests it acts as a functional readout of an individual's underlying "immunological setpoint." A healthy, diverse, and resilient microbiome, rich in anti-inflammatory species, may be able to withstand the insult of SARS-CoV-2 infection, helping to orchestrate a balanced immune response and facilitate recovery. Conversely, a pre-existing or acutely-induced dysbiosis may represent a state of lower resilience. When stressed by the virus, this fragile ecosystem collapses, failing to regulate the immune response and allowing the cascade of events leading to Long COVID—including the establishment of a viral reservoir and chronic inflammation—to proceed unchecked. In this view, Long COVID is not just a consequence of the virus itself, but a failure of host-microbe homeostasis.
The specific microbial signature of the Long COVID gut has been characterized across numerous studies. It consistently features a continued depletion of beneficial butyrate-producing bacteria, such as Faecalibacterium prausnitzii and various species of Bifidobacterium and Dorea.18 Concurrently, there is a persistent enrichment of pro-inflammatory or opportunistic genera, including Actinomyces, Collinsella, Bacteroides, Flavonifractor, Leuconostoc, and Granulicatella.18
This dysbiotic signature is not monolithic; specific microbial patterns are correlated with distinct symptom clusters of Long COVID. For example, one study found that persistent respiratory symptoms were strongly associated with an enrichment of opportunistic bacteria like Streptococcus species, while neurological symptoms and fatigue were linked to pathogens such as Clostridium innocuum.18 Multi-omics analyses have shown that microbiome data is particularly effective at predicting the presence of gastrointestinal, emotional, and sleep disturbances.27 This suggests that different microbial imbalances may drive pathology through different pathways, contributing to the heterogeneous clinical presentation of Long COVID.
The disruption also extends beyond the bacterial domain to the gut mycobiome (the community of fungi). A two-year longitudinal study tracking COVID-19 patients found long-term shifts in gut fungi composition. The enrichment of certain yeast genera, like Hanseniaspora and Saturnispora, was significantly correlated with persistently impaired pulmonary function two years after infection, providing a clear example of the gut-lung axis at play in the context of Long COVID.28
Table 2: Comparative Gut Microbial Signatures in Acute COVID-19 and Long COVID
This table contrasts the key features of gut dysbiosis observed during the acute phase of infection with the chronic dysbiotic state characteristic of Long COVID.
Part III: A Unified Pathophysiological Model of Gut-Mediated Long COVID
The evidence from mechanistic and observational studies provides the essential components of a complex puzzle. This section synthesizes these components into a cohesive, multi-step pathophysiological model that explains how events initiated in the gut can precipitate the systemic, multi-organ disease known as Long COVID. This model posits a central cascade: persistent viral infection and dysbiosis lead to intestinal barrier failure, which in turn allows the translocation of inflammatory triggers into the bloodstream, driving chronic systemic inflammation and metabolic disruption.
Section 3.1: The "Leaky Gut" Nexus: Zonulin, Microbial Translocation, and Systemic Inflammation
The lynchpin connecting localized gut pathology to systemic illness is the failure of the intestinal barrier, a condition colloquially known as "leaky gut." This breakdown allows molecules that should be contained within the gut lumen to enter the bloodstream, where they trigger a chronic, low-grade inflammatory response throughout the body.
At the heart of this barrier failure is a protein called zonulin. Zonulin is a physiological modulator of the tight junctions, the protein complexes that seal the space between adjacent intestinal epithelial cells, forming a physical barrier.29 When zonulin is released, it binds to receptors on these cells, triggering a signaling cascade that causes the tight junctions to temporarily open. While this is a normal physiological process, chronic overexpression of zonulin leads to a state of pathologically increased intestinal permeability.29
A growing body of evidence implicates zonulin as a key culprit in Long COVID. Multiple human studies have found that individuals with Long COVID have significantly higher circulating levels of zonulin compared to both people who recovered fully from COVID-19 and uninfected controls.31 The association is robust; one large study found that every one-unit increase in a patient's zonulin level was associated with a 44% higher odds of having PASC.32 While the precise trigger for this elevated zonulin is still under investigation, a leading hypothesis is that the persistent presence of SARS-CoV-2 spike protein in the gut mucosa directly induces its overexpression.31 This mechanism is strongly supported by parallel findings in Multisystem Inflammatory Syndrome in Children (MIS-C), a severe post-COVID condition where gut viral persistence has been shown to drive zonulin release and subsequent barrier failure.33
This zonulin-mediated breach of the gut barrier has profound consequences. It opens the floodgates for a host of inflammatory molecules to translocate from the gut into the systemic circulation. These include:
SARS-CoV-2 Antigens: The persistent viral reservoir in the gut can now continuously leak viral components, most notably the spike protein, into the bloodstream. The detection of these antigens in the blood of Long COVID patients months after infection provides a direct source of chronic antigenic stimulation for the immune system.19
Microbial Products: The barrier breach also allows for the translocation of bacterial components. This includes potent pro-inflammatory molecules like lipopolysaccharide (LPS), a component of the outer membrane of Gram-negative bacteria, and other pathogen-associated molecular patterns (PAMPs).30 Indeed, studies have found elevated levels of markers of microbial translocation in the blood of Long COVID patients.32
This constant, low-level seeding of the bloodstream with gut-derived viral and microbial products is believed to be a primary driver of the chronic, systemic inflammation and immune dysregulation that are hallmarks of Long COVID, providing a direct mechanistic link between the gut and the patient's systemic symptoms.32
Section 3.2: Systemic Consequences of Altered Microbial Metabolism
The gut microbiome is a vast metabolic organ, converting dietary components and host products into a myriad of bioactive molecules that influence host physiology. The profound dysbiosis seen in Long COVID results in a dysfunctional metabolic output, leading to a systemic deficit of beneficial compounds and a surplus of harmful ones, with far-reaching consequences for immunity, energy metabolism, and neurological function.
The most consistently reported metabolic defect is a depletion of the short-chain fatty acid (SCFA) butyrate.18 This is a direct consequence of the loss of major butyrate-producing bacteria like Faecalibacterium prausnitzii.18 Butyrate is the preferred energy source for colonocytes, and its absence compromises the health and integrity of the intestinal lining itself, further contributing to barrier dysfunction. However, its effects are not just local. Butyrate that enters the circulation has potent systemic anti-inflammatory properties, including the ability to promote the development of regulatory T cells (T-regs), which are crucial for suppressing excessive immune responses.35 It also plays a key role in supporting mitochondrial function and energy metabolism.36 The chronic deficit of butyrate in Long COVID patients is therefore thought to contribute directly to the state of persistent inflammation and the profound fatigue and post-exertional malaise that are characteristic of the condition.
Another critical pathway disrupted by gut dysbiosis in Long COVID is the metabolism of the essential amino acid tryptophan. Under normal conditions, gut microbes play a key role in tryptophan metabolism, which is the precursor for the neurotransmitter serotonin. Approximately 90% of the body's serotonin is produced in the gut, where it acts as a critical signaling molecule.20 In Long COVID, a multi-pronged assault disrupts this pathway. First, the persistent inflammation in the gut wall, driven by the viral reservoir, impairs the absorption of dietary tryptophan.20 Second, the dysbiotic microbiome has a reduced capacity to produce beneficial tryptophan metabolites like indolepropionic acid (IPA), which have neuroprotective and anti-inflammatory effects.37 The net result is a systemic depletion of both tryptophan and serotonin. This has devastating consequences for the microbiota-gut-brain axis, as serotonin is a vital regulator of the vagus nerve, the primary bidirectional communication highway between the gut and the brain.20 The resulting vagus nerve dysfunction is a leading hypothesis to explain many of the neurological symptoms of Long COVID, including memory loss, difficulty concentrating, and "brain fog".20 This mechanism is supported by human metabolomics studies that confirm Long COVID patients have lower serum levels of tryptophan and IPA, and higher levels of inflammatory tryptophan metabolites.37
Beyond these specific pathways, broader metabolomic profiling of plasma from Long COVID patients reveals a state of profound and persistent metabolic chaos. These studies show widespread dysregulation of lipid metabolism (including sphingolipids and glycerophospholipids), impaired energy metabolism (evidenced by the accumulation of lactic acid), and a systemic redox state imbalance.38 Remarkably, these metabolic disturbances have been shown to persist for at least two years after the initial infection, underscoring the deep and long-lasting systemic impact of the initial gut-centered pathology.38
Section 3.3: The Microbiota-Gut-Brain Axis in Long COVID
The debilitating neurological and cognitive symptoms of Long COVID—often termed "neuro-PASC"—are among the most distressing and difficult to treat. These symptoms, which include "brain fog," memory impairment, difficulty concentrating, mood disturbances, and severe fatigue, are not isolated phenomena occurring solely within the central nervous system. Instead, the evidence strongly indicates that they are a direct consequence of the gut pathology, communicated to the brain via the intricate network of the microbiota-gut-brain axis.2
The mechanisms by which the disrupted gut environment impacts the brain in Long COVID are multifactorial and synergistic.2 They include:
Direct Neuroinflammation: The systemic inflammation fueled by the "leaky gut" is a key driver. Pro-inflammatory cytokines and activated immune cells that originate in response to the gut reservoir and microbial translocation can cross the normally restrictive blood-brain barrier. Once in the central nervous system, they can activate resident immune cells like microglia, creating a state of chronic neuroinflammation that impairs neuronal function and cognitive processes.2
Vagus Nerve Dysfunction: As detailed previously, the disruption of gut-derived serotonin production directly impairs the signaling of the vagus nerve.20 This disrupts the primary neural communication pathway from the gut to the brain, which is critical for regulating mood, memory, and autonomic function. This pathway is a prime candidate for explaining the rapid onset of cognitive and mood symptoms in Long COVID.20
Altered Microbial Metabolites: The brain is highly sensitive to the metabolic milieu of the body. The chronic deficit of beneficial microbial metabolites like butyrate, which supports neuronal health, coupled with an increase in potentially neurotoxic byproducts from a dysbiotic microbiome, can directly impact brain function. These metabolic shifts can affect critical processes like adult neurogenesis (the creation of new neurons) and myelination (the maintenance of the protective sheath around nerve fibers), contributing to long-term cognitive decline.2
The validity of this gut-brain model for Long COVID is further strengthened by the striking parallels observed with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS), another complex, post-viral illness characterized by similar neurological symptoms. Research into ME/CFS has also identified profound gut dysbiosis, altered microbial metabolism (including butyrate and tryptophan deficits), and immune dysregulation as core features.27 The significant overlap in both the clinical presentation and the underlying gut-centric pathophysiology between Long COVID and ME/CFS strongly suggests they may share a common mechanistic basis of post-viral, gut-mediated, neuro-immune disruption.35
Part IV: Clinical and Therapeutic Implications
The comprehensive, gut-centric model of Long COVID pathogenesis is not merely an academic exercise. It provides a powerful, evidence-based framework for developing novel clinical strategies. If a persistent viral reservoir, chronic gut dysbiosis, and barrier failure are the upstream drivers of the disease, then this opens a new frontier for diagnostics that can objectively identify these pathologies and for therapeutics that can directly target them. This section explores the emerging clinical and therapeutic implications of this gut-focused paradigm.
Section 4.1: Microbiome-Based Diagnostics and Prognostics
A major challenge in managing Long COVID has been the lack of objective, reliable biomarkers to diagnose the condition and predict its course. The distinct microbial and metabolic signatures associated with Long COVID offer a promising solution to this problem, paving the way for a new generation of diagnostic and prognostic tools.
The predictive power of the acute-phase gut microbiome represents a significant opportunity for early intervention. As established, machine learning models can use gut microbiome data from the initial phase of COVID-19 to identify, with high accuracy, which individuals are at greatest risk of developing Long COVID.21 In the future, this could translate into a prognostic test administered during or shortly after acute infection. Such a test would allow clinicians to triage at-risk patients for pre-emptive therapies or closer monitoring, potentially preventing the full establishment of the chronic disease state.
For patients who already have established Long COVID, specific microbial signatures could serve as diagnostic biomarkers. Studies have identified a panel of bacterial genera—notably Leuconostoc, Actinomyces, and Granulicatella—that are consistently enriched in Long COVID patients and can accurately distinguish them from healthy controls and those who have recovered from COVID-19 without sequelae.25 These bacteria could form the basis of a non-invasive, stool-based diagnostic test to provide objective evidence for a Long COVID diagnosis, which is currently based on subjective symptom reporting.
The future of diagnostics, however, likely lies in integrated, multi-omics approaches. The complexity of Long COVID suggests that a single biomarker may be insufficient. Advanced artificial intelligence platforms, such as the BioMapAI tool used in ME/CFS research, demonstrate the power of combining data from multiple biological layers.27 By integrating gut metagenomics (what microbes are present), plasma metabolomics (what those microbes are producing), and immune profiling (how the host is responding), these systems can create a highly detailed and accurate "map" of the disease state. This approach has achieved approximately 90% accuracy in classifying patients with ME/CFS, a close proxy for Long COVID.27 Adopting such a systems-biology approach could lead to diagnostic panels that not only confirm a Long COVID diagnosis but also stratify patients into different endotypes based on their specific underlying pathophysiology (e.g., "inflammatory-dominant," "metabolic-dominant," "neuro-dominant"), allowing for more precise and personalized therapeutic interventions.
Section 4.2: Therapeutic Interventions Targeting the Gut-Microbiome Axis
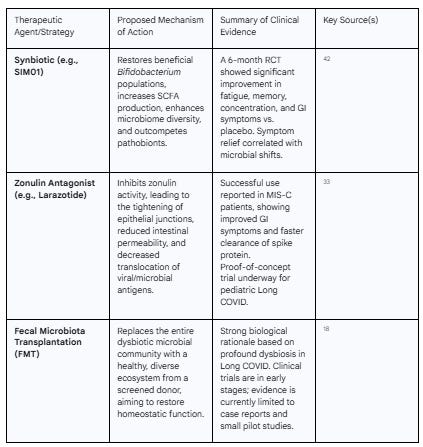
If the gut is a central driver of Long COVID, then therapies aimed at restoring a healthy microbiome and healing the intestinal barrier are a logical and highly promising treatment strategy. Several such interventions are now being investigated in clinical trials, with encouraging initial results that validate the gut as a key therapeutic target.
One of the most advanced strategies involves the use of probiotics, prebiotics, or synbiotics (a combination of both) to actively modulate the gut microbiome. A landmark randomized, placebo-controlled trial investigated the effects of a synbiotic formulation called SIM01, which contains specific strains of Bifidobacterium and prebiotic fibers, in Long COVID patients.42 After six months of treatment, the group receiving SIM01 reported significantly greater improvement in multiple core Long COVID symptoms—including fatigue, memory loss, difficulty concentrating, and gastrointestinal upset—compared to the placebo group.42 Crucially, this clinical improvement was directly correlated with positive changes in the gut microbiome: the SIM01 group showed increased microbial diversity and a specific increase in the abundance of the beneficial Bifidobacterium species that were part of the formulation.42 This trial provides direct, high-quality evidence that therapeutically modulating the gut microbiome can lead to tangible clinical benefits in Long COVID patients.
A more radical approach to microbiome modulation is Fecal Microbiota Transplantation (FMT), which aims to completely reset a patient's dysbiotic microbiome by introducing a healthy, diverse microbial community from a screened donor. While large-scale, controlled trials in Long COVID are still pending, the strong rationale for its use, based on the profound dysbiosis observed in patients, has made it an area of intense research interest. Case reports and small pilot studies are emerging, and FMT is considered a powerful potential therapy for refractory cases.18
Beyond modulating the microbes themselves, another novel strategy is to directly target the compromised gut barrier. As elevated zonulin and the resulting "leaky gut" are central to the disease model, drugs that can inhibit zonulin and tighten the intestinal junctions are a compelling therapeutic option. The drug larazotide is a zonulin antagonist specifically designed for this purpose.29 A proof-of-concept clinical trial is currently underway to test the efficacy of larazotide in children with Long COVID.33 The rationale for this trial is built on the successful use of larazotide in a small number of children with the related condition MIS-C. In those cases, treatment with larazotide led to faster improvement of GI symptoms and, remarkably, accelerated the clearance of SARS-CoV-2 spike protein from the blood.33 This suggests that by "plugging the leak" in the gut, larazotide may be able to cut off the supply of inflammatory triggers to the rest of the body. This represents a fundamentally new therapeutic approach that targets the host's barrier integrity rather than the microbes themselves.
Table 3: Emerging Gut-Targeted Therapies for Long COVID
This table summarizes the primary therapeutic strategies being investigated to target the gut-centric pathophysiology of Long COVID.
Conclusion: Future Research Trajectories and the Path to Precision Medicine
The convergence of mechanistic and clinical evidence has solidified a gut-centric model as a leading explanation for the pathophysiology of Long COVID. This model provides a coherent narrative that links an initial viral infection to the chronic, multisystemic symptoms that afflict millions worldwide. The cascade—from direct enteric infection and the establishment of a persistent viral reservoir, to chronic gut dysbiosis, zonulin-mediated barrier failure, and the subsequent systemic inflammation and metabolic disruption driven by microbial translocation and dysfunctional metabolite production—offers a robust framework for understanding this complex post-viral syndrome.
However, this understanding is the beginning, not the end, of the scientific journey. The path forward requires a concerted research effort focused on several key areas. First, there is a critical need for larger, more geographically diverse, and longitudinal multi-omics studies. These studies must track patients from the acute phase of infection through the development and progression of Long COVID, integrating metagenomics, metabolomics, proteomics, and detailed immunological profiling. This will be essential for validating the diagnostic and prognostic biomarkers identified in initial cohorts and for further dissecting the specific microbial pathways that are linked to different symptom clusters.
Second, the promising initial results from gut-targeted therapeutic trials must be followed by more rigorous, large-scale, placebo-controlled clinical trials. The efficacy of different synbiotic formulations, the optimal timing and application of FMT, and the therapeutic potential of barrier-healing drugs like larazotide must be systematically evaluated. These trials should incorporate multi-omics analyses as secondary endpoints to confirm that clinical improvement is indeed accompanied by the intended modulation of the microbiome and host response.
Ultimately, the goal is to move beyond a one-size-fits-all approach and toward a future of precision medicine for Long COVID. The heterogeneity of the syndrome strongly suggests that different patients may have different primary drivers of their illness. By using advanced diagnostic platforms to stratify patients based on their unique gut microbiome and metabolomic profiles, it may become possible to guide the selection of the most effective personalized interventions. A patient with severe butyrate deficiency might benefit most from a targeted synbiotic, while a patient with extremely high zonulin levels might be a prime candidate for a barrier-healing drug. For those with the most profoundly disrupted and resiliently dysbiotic ecosystems, a full microbial reset with FMT may be the most appropriate course of action. By continuing to unravel the intricate connections between SARS-CoV-2, the gut microbiome, and the host, the scientific and medical communities can pave the way for evidence-based strategies that can finally offer relief and recovery to those living with the long shadow of COVID-19.
Acknowledgement
I acknowledge the assistance of Gemini AI in the preparation of the subject research plan, the execution of the research, and the preparation of this report.
Works cited
Translating animal models of SARS-CoV-2 infection to vascular, neurological and gastrointestinal manifestations of COVID-19 - Company of Biologists Journals, accessed July 31, 2025, https://journals.biologists.com/dmm/article/18/9/dmm052086/367629/Translating-animal-models-of-SARS-CoV-2-infection
Role of the microbiota-gut-brain axis in postacute COVID syndrome ..., accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC10042594/
Gut microbiota in COVID-19: key microbial changes, potential ..., accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC9589856/
Impact of SARS-CoV-2 infection on respiratory and gut microbiome stability: a metagenomic investigation in long-term-hospitalized COVID-19 patients - PubMed Central, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC11561083/
Mechanisms Leading to Gut Dysbiosis in COVID-19: Current ..., accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC9505288/
Variable susceptibility of intestinal organoid–derived monolayers to ..., accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC9004766/
SARS-CoV-2 Coronavirus Can Infect Gut Enterocytes | Sci.News, accessed July 31, 2025, https://www.sci.news/medicine/sars-cov-2-coronavirus-gut-enterocytes-08392.html
Organoids demonstrate gut infection by SARS-CoV-2 - PubMed, accessed July 31, 2025, https://pubmed.ncbi.nlm.nih.gov/32427981/
Gut microbiome dysbiosis during COVID-19 is associated with increased risk for bacteremia and microbial translocation - PubMed Central, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC8902880/
Animal models for SARS‐CoV‐2 infection and pathology - PMC, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC8662225/
Animal Models to Test SARS-CoV-2 Vaccines: Which Ones Are in Use and Future Expectations - MDPI, accessed July 31, 2025, https://www.mdpi.com/2076-0817/12/1/20
Animal Models for COVID-19: Hamsters, Mouse, Ferret, Mink, Tree Shrew, and Non-human Primates - Frontiers, accessed July 31, 2025, https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2021.626553/full
Pathogenesis and virulence of coronavirus disease: Comparative pathology of animal models for COVID-19 - PMC - PubMed Central, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC10878030/
Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19, accessed July 31, 2025, https://gut.bmj.com/content/70/2/276
Altered gut microbiota patterns in COVID-19: Markers for inflammation and disease severity - PubMed Central, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC9280735/
Microbiome dysbiosis in SARS-CoV-2 infection: implication for pathophysiology and management strategies of COVID-19 - PubMed Central, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC12052750/
Attenuated replication and damaging effects of SARS-CoV-2 Omicron variants in an intestinal epithelial barrier model | bioRxiv, accessed July 31, 2025, https://www.biorxiv.org/content/10.1101/2024.02.28.582510v1.full-text
Covid Alteration of the Gut Microbiome: A Factor in Long Covid? - Global Virus Network, accessed July 31, 2025, https://gvn.org/covid-alteration-of-the-gut-microbiome-a-factor-in-long-covid/
COVID-19 Virus Can Stay in the Body More Than a Year after ..., accessed July 31, 2025, https://www.ucsf.edu/news/2024/03/427241/covid-19-virus-can-stay-body-more-year-after-infection
Viral persistence and serotonin reduction can cause long COVID ..., accessed July 31, 2025, https://www.eurekalert.org/news-releases/1004844
Gut Microbiome Signatures During Acute Infection Predict Long ..., accessed July 31, 2025, https://www.biorxiv.org/content/10.1101/2024.12.10.626852v1.full
Gut Microbiome During COVID-19 Infection Predicts Long COVID Risk - EMJ, accessed July 31, 2025, https://www.emjreviews.com/respiratory/news/gut-microbiome-during-covid-19-infection-predicts-long-covid-risk/
Full article: Robust cross-cohort gut microbiome associations with COVID-19 severity, accessed July 31, 2025, https://www.tandfonline.com/doi/full/10.1080/19490976.2023.2242615
Full article: Long COVID and gut microbiome: insights into ..., accessed July 31, 2025, https://www.tandfonline.com/doi/full/10.1080/19490976.2025.2457495
Gut Microbial Signatures in Long COVID: Potential Biomarkers and Therapeutic Targets, accessed July 31, 2025, https://pubmed.ncbi.nlm.nih.gov/40450635/
Gut Microbial Signatures in Long COVID: Potential Biomarkers and ..., accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC12271030/
Gut microbiome may predict invisible chronic fatigue syndrome and ..., accessed July 31, 2025, https://www.jax.org/news-and-insights/2025/july/gut-microbiome-may-predict-invisible-chronic-fatigue-syndrome-and-long-covid
Longitudinal alterations of gut mycobiota during 2 years after COVID-19 and its correlation with pulmonary sequela, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC12210925/
Zonulin as a Potential Therapeutic Target in Microbiota-Gut-Brain Axis Disorders: Encouraging Results and Emerging Questions - PubMed Central, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC10139156/
Evaluation of serum zonulin and occludin levels in obsessive-compulsive disorder and the effect of major depressive disorder comorbidity - Frontiers, accessed July 31, 2025, https://www.frontiersin.org/journals/psychiatry/articles/10.3389/fpsyt.2024.1395235/full
Zonulin, a marker of gut permeability, is associated with mortality in a cohort of hospitalised peruvian COVID-19 patients, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC9485714/
Increase in gut permeability and oxidized ldl is associated ... - Frontiers, accessed July 31, 2025, https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2023.1182544/full
SARS-CoV-2 gut persistence, intestinal permeability, and spike ..., accessed July 31, 2025, https://polybio.org/projects/sars-cov-2-gut-persistence-intestinal-permeability-and-spike-protein-in-pediatric-long-covid-mis-c/
Loss of gut epithelial barrier responsible for COVID-19-related MIS-C in children, suggests study - News-Medical.net, accessed July 31, 2025, https://www.news-medical.net/news/20210528/Loss-of-gut-epithelial-barrier-responsible-for-COVID-19-related-MIS-C-in-children-suggests-study.aspx
Gut bacteria could help diagnose long Covid and chronic fatigue syndrome, researchers find, accessed July 31, 2025, https://www.independent.co.uk/news/health/gut-bacteria-long-covid-chronic-fatigue-study-b2795311.html
Why Butyrate Matters in ME/CFS and Long COVID - YouTube, accessed July 31, 2025,
Full article: Dysrupted microbial tryptophan metabolism associates with SARS-CoV-2 acute inflammatory responses and long COVID - Taylor & Francis Online, accessed July 31, 2025, https://www.tandfonline.com/doi/full/10.1080/19490976.2024.2429754
The plasma metabolome of long COVID patients two years after infection - ResearchGate, accessed July 31, 2025, https://www.researchgate.net/publication/372807353_The_plasma_metabolome_of_long_COVID_patients_two_years_after_infection
Proteomic and metabolomic profiling of plasma uncovers immune responses in patients with Long COVID-19 - PMC, accessed July 31, 2025, https://pmc.ncbi.nlm.nih.gov/articles/PMC11718655/
Gut microbiome may predict “invisible” chronic fatigue syndrome and long COVID, accessed July 31, 2025, https://www.eurekalert.org/news-releases/1092282
Your Gut May Be the Key to Chronic Fatigue, Long COVID - Neuroscience News, accessed July 31, 2025, https://neurosciencenews.com/chronic-fatigue-long-covid-microbiome-29530/
Experimental drug that alters gut microbiome shows promise for ..., accessed July 31, 2025, https://www.cidrap.umn.edu/covid-19/experimental-drug-alters-gut-microbiome-shows-promise-long-covid-relief